Class
FragmentStore
Multi-Container to store contigs, reads and a multiple alignment between them.
Include Headers
seqan/store.h
Parameters
The specializing type. Default: | |
The configuration struct. Default: |
Typedefs
| Type of the alignedReadStore member. | |
| Type of the alignedReadTagStore member. | |
| Type of the alignQualityStore member. | |
| Type of the annotationKeyStore member. | |
| Type of the annotationNameStore member. | |
| Type of the annotationStore member. | |
| Type of the annotationTypeStore member. | |
| Type of the contigFileStore member. | |
| Type of the contigNameStore member. | |
| Type of the contigStore member. | |
| Type of the libraryNameStore member. | |
| Type of the libraryStore member. | |
| Type of the matePairNameStore member. | |
| Type of the matePairStore member. | |
| Type of the readNameStore member. | |
| Type of the readSeqStore member. | |
| Type of the readStore member. |
Data Members
| String that stores | |
| StringSet that maps from | |
| String that maps from | |
| StringSet that maps from | |
| StringSet that maps from | |
| String that maps from | |
| StringSet that maps from | |
| String that maps from | |
| StringSet that maps from | |
| String that maps from | |
| StringSet that maps from | |
| String that maps from | |
| StringSet that maps from | |
| String that maps from | |
| StringSet that maps from | |
| StringSet that maps from | |
| String that maps from |
Functions
| Appends an aligned read entry to a fragment store. | |
| Appends two paired-end reads to a fragment store. | |
| Appends a read to a fragment store. | |
| The begin of a container. | |
| Calculates a string with insert sizes for each pair match. | |
| Calculates a string that maps the | |
| Removes all contigs from a fragment store. | |
| Removes all reads from a fragment store. | |
| Removes invalid aligned reads and rename | |
| Renames | |
| Converts all matches to a multiple global alignment in gap-space. | |
| Converts pair-wise alignments to a multiple global alignment. | |
| The end of a container. | |
| Get the "clear" range of a read alignment. | |
| Returns the mate number of read for a given | |
| Returns the read with the given | |
| Calculates a visible layout of aligned reads. | |
| Manually loads a contig sequence. | |
| Loads contigs into fragment store. | |
| Loads reads into fragment store. | |
| Locks a contig sequence from being removed. | |
| Locks all contig sequences from being removed. | |
| Prints a window of the visible layout of reads into a outstream. | |
| Loads records from a file. | |
| Removes a previous contig lock and clears sequence no further lock exist. | |
| Removes a previous lock for all contigs and clears sequences without lock. | |
| Removes a previous contig lock. | |
| Removes a previous lock for all contigs. | |
| Saves records to a file. | |
| Write contigs from fragment store into file. |
Examples
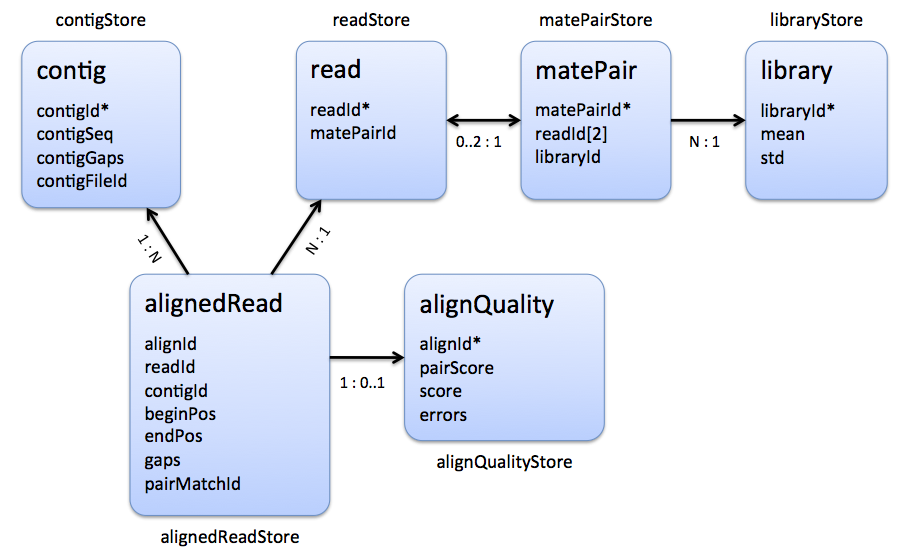 |
| Stores that are involved in the representation of a multiple read alignment. |
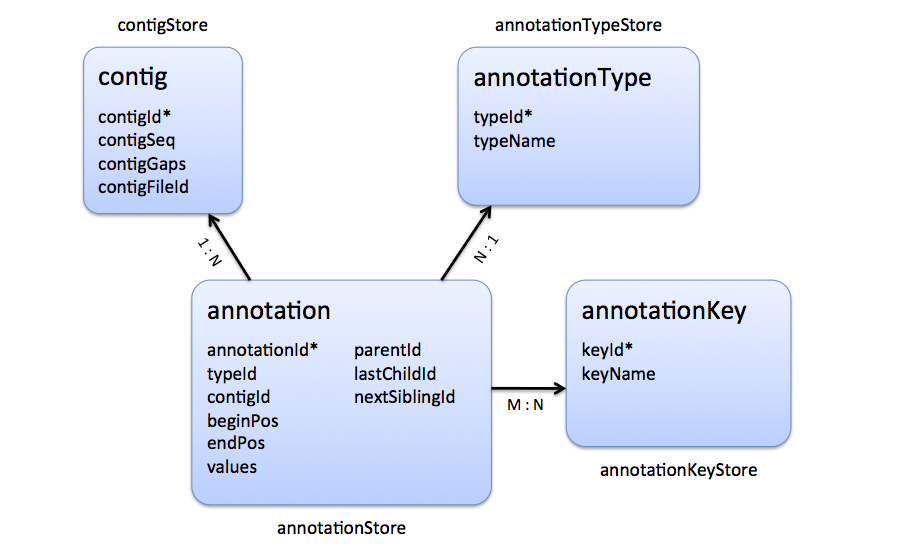 |
| Stores that are involved in the representation of a genome alignment. |
SeqAn - Sequence Analysis Library - www.seqan.de